Monitoring of Venturia inaequalis virulences
Establishing a network of orchards
of differential hosts
Selection of the differential hosts
A first set of differential hosts used to assign specific Venturia
inaequalis isolates to a certain race, developed
over time from the works of Shay and Williams (1956), Shay et al.
(1962), Williams and Brown
(1968), Parisi et al. (1993), Bénaouf and Parisi (2000) and
Bus et al. (2005) (Table
1). The more the apple-apple scab interaction
was studied, more evident become the problem of the current nomenclature
to assign to a race an isolate able to grow on two differential hosts.
To solve these conflicts Bus et al. (2008) proposed a new
nomenclature which is better suited to describe
the increasing complexities of combinations of genes (from scab and
the apple) involved in both
race-specific and race-nonspecific interactions. In this attempt
the “old” set of differential hosts was revisited. Previously
not considered accessions carrying R-genes that could be interesting
for breeding purposes have been included and differential host of
the “old” set known to carry more than an R-gene have
been substituted with selections carrying only one of these known
R-genes. In order to coordinate the genetic studies of the Venturia
inaequalis-Malus pathosystem at laboratory scale with the monitoring of apple scab
virulences in the field, the same differential hosts proposed by
Bus et al. (2008) with few additions have been chosen for the monitoring (Table
2).
This set of differential host reflects the knowledge of the V. inaequalis-Malus pathosystem of June 2008. The set will be up-dated each time new apple scab R-genes are discovered and sufficient plant material is available. Currently Rvi16 discovered in MIS op 93.051 G07-098 (Bus et al. 2010) and Rvi17 of Antonovka APF22 (Dunneman and Egerer 2010) are missing in the set. As soon plant material of these genotypes will be available, they will be added to the set.
Verification of the genotypes selected as differential hosts
All the selected genotypes have been molecularly checked. The genotypes
and reference DNAs (requested to different institutes) have been
analyzed with 14 SSRs selected by ECPGR to molecularly characterize
apples. The fingerprints have been compared to verify if the genoytpes
available at ACW are “true-to-type”. The second check
consisted in testing the genotypes with the molecular
markers associated to their R-genes (if markers were available) to confirm the presence
of the expected R-genes. Plants of genotypes found “not-true-to-type” have
been replaced.
Table 1: Set of “old” differential
hosts (adapted from Gessler et al. 2006)
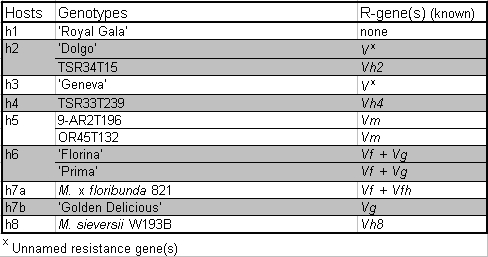
Table 2: New set of differential hosts (adapted from Bus et al.
2008)